
Spatial Transcriptomics
Single-cell RNA sequencing allows us to understand the gene expression profiles of individual cells within a heterogeneous population, providing insights into cell diversity, differentiation, and other biological processes. Spatial transcriptomics builds upon this foundation by visualizing gene expression patterns within the tissue environment, depicting the relative positions of cells in tissue samples and their interrelationships.
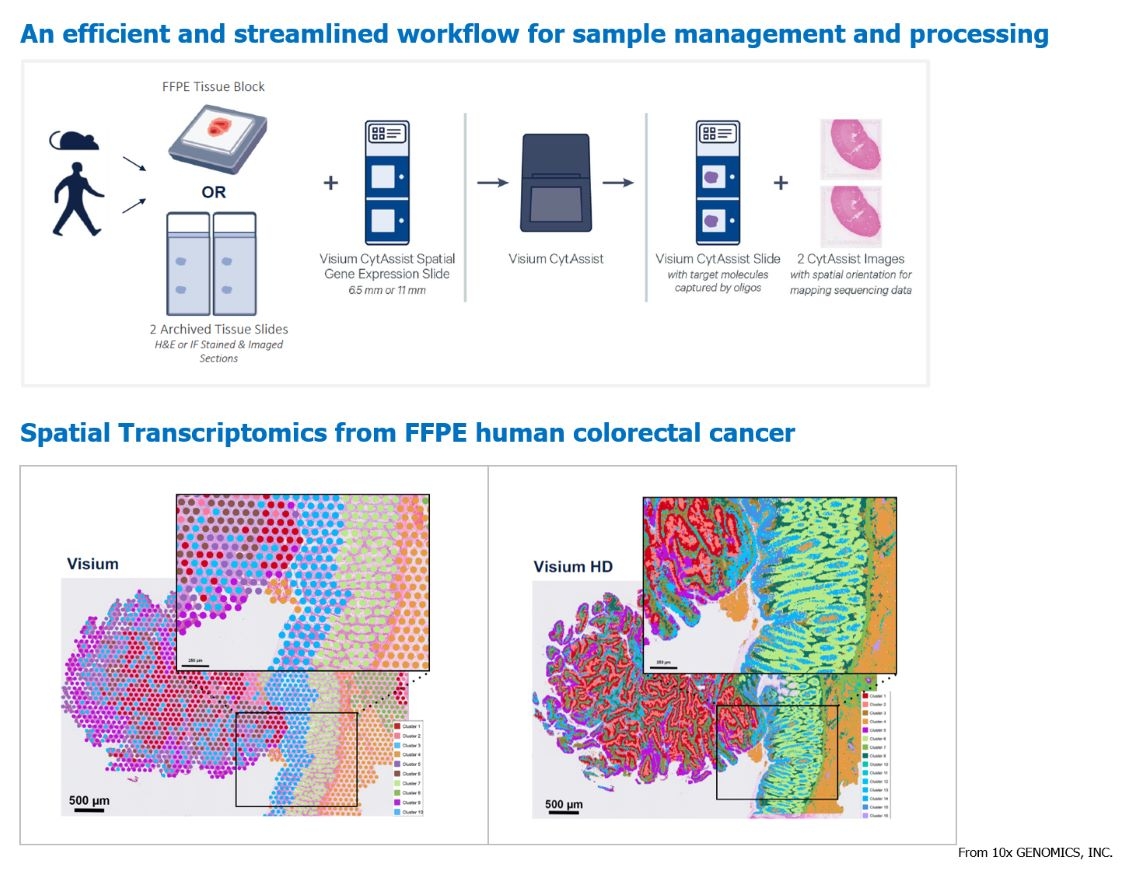
Spatial transcriptomics is achieved using the 10X Genomics Visium platform, which integrates microscopy, chips, and sequencing technologies. It involves the use of spatially barcoded probes on intact fresh-frozen (FF) or formalin-fixed paraffin-embedded (FFPE) tissue sections. These probes identify captured targets on slides, enabling the extraction of transcriptomic data from cells at different locations. This approach combines histology with gene expression analysis and utilizes specific data analysis methods to visualize gene expression and morphological data. It provides immunological maps, insights into tumor microenvironments, and spatial mapping of different tissues, enhancing our understanding of biological processes and disease pathology. 10X Genomics' original Visium provided spatial resolution covering 1-10 cells per spot. In this year, they launched Visium HD, a higher-resolution assay kit that achieves sequencing resolution down to the level of single cells. This advancement in spatial transcriptomics offers clearer insights.
Sample types: Formalin-fixed paraffin-embedded tissue blocks, H&E-stained or immunofluorescence-stained tissue sections, and their corresponding microscopic imaging files.
Strain Identification
Advantages of Strain Identification Service :
1. Genetic identification methods analyze bacterial 16S and fungal ITS, capable of identifying at the "species" level.
2. Both agar plates and liquid cultures of individual strains are applicable. Genomic DNA of microorganisms can also be submitted for identification.
3. Reports are generated within 5 working days upon sample receipt.
4. A dedicated data management system is available for customers to download files.