
Strain Identification
Service advantages :
- ISO/IEC 17025 testing laboratory with professional quality you can trust.
- Genetic identification methods analyze bacterial 16S and fungal ITS, capable of identifying at the "species" level.
- Reports are generated within 5 working days upon sample receipt.
Sample conditions :
- Bacteria and fungi can both be identified.
- Both agar plates and liquid cultures of individual strains are applicable.
- Genomic DNA of microorganisms can also be submitted for identification.
- Please send the samples refrigerated at 4°C.
Description :
- The sample genomic DNA is amplified using species-specific primer pairs for Strain Identification, followed by sequencing reactions. Comparative analysis is then performed on the sequenced results.
- If you would like to learn more about the testing scope of Strain Identification conducted by the Genetic Lab. Of TRI-I BIOTECH, please click on the TAF official link below for further information.
Sample type :(Please choose the options :Bacteria, Fungus, or Genomic DNA)
- Single strain of Plate (or Broth).
- Genomic DNA.
Acceptance criteria :
- The absorbance value A260/A280 ratio must be between 1.6-2.0.
- If the concentration of DNA is lower than 10 ng/μl (including 10 ng/μl) and there is no PCR product, re-extraction is required.
Reporting of results :
- Exclusive test report format for Strain Identification.
- Provide the original wave peak diagram after sequencing.
- Performing analysis using professional bioinformatics software and providing results and files for sequence alignment.
★If you would like to entrust Strain Identification service, please click on the Laboratory Login. After logging in, you can proceed with the order.
★If you need to download the sample testing service order form for Strain Identification, please click on the DOWNLOAD button.
Information of Strain Identification

Microbial Community Analysis Service
Multi-source Microbial Community Analysis Service (16S/ITS Metagenomics)
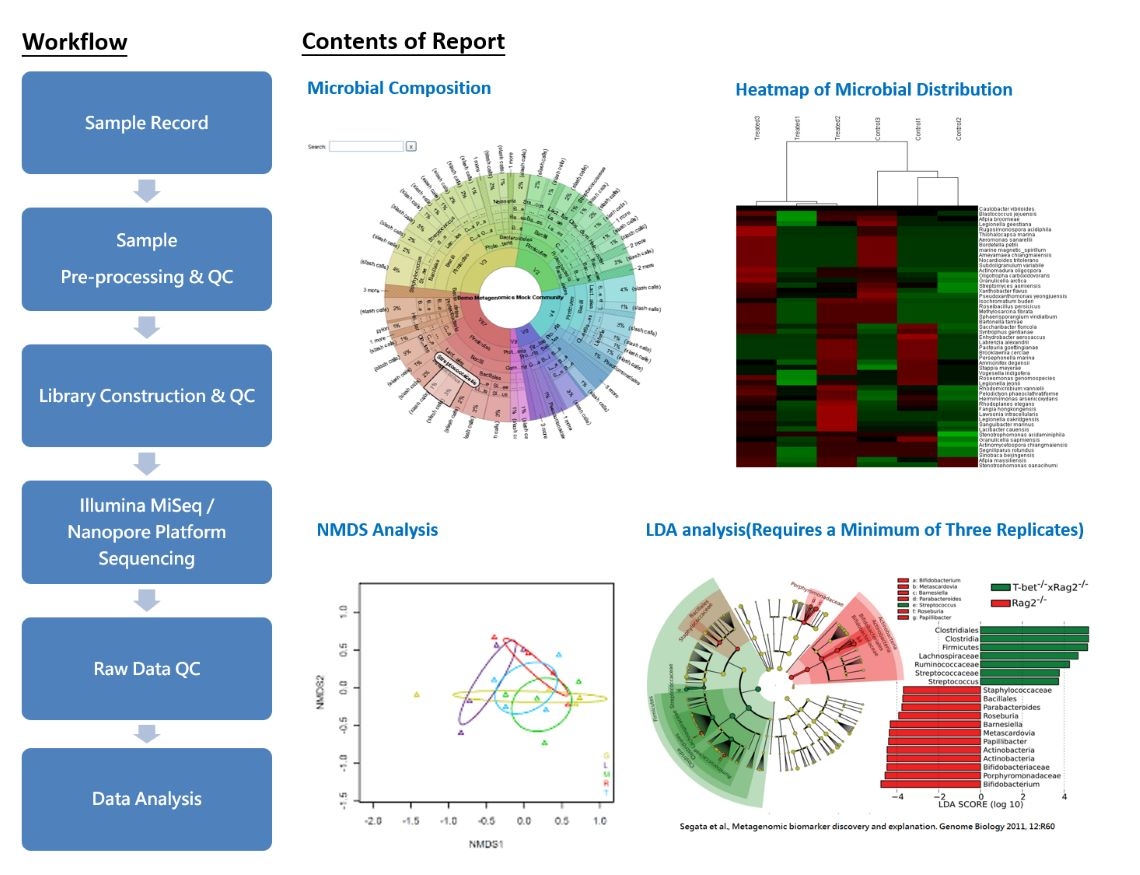
Metagenomics, also known as Environmental Genomics, is the scientific study of an entire microbial community. It involves extracting DNA from environmental samples containing microorganisms, sequencing the DNA, and then conducting analyses such as species identification, phylogenetic assessments, gene function studies, and phenotype correlations. The aim is to gain insights into the diversity of microorganisms present in the environment.
The 16S rRNA gene is present in the ribosomes of all bacteria and exhibits highly conserved sequences as well as diverse features among different bacterial species. Consequently, it serves as a valuable marker for bacterial classification and identification. 16S metagenomics primarily focuses on the sequencing of the 16S rRNA gene sequences of bacteria. This involves amplifying rRNA fragments through PCR, constructing paired-end libraries, and utilizing the Illumina MiSeq platform for sequencing. Subsequently, the obtained sequences are analyzed to determine the bacterial taxa and their distribution proportions in each sample based on sequence variations and abundance.
In summary, the application of metagenomics provides a comprehensive and in-depth understanding of microbial community structure and function. This approach is crucial for ecological studies, environmental science, and research related to microbial health.
Sample Requirements:
Sample Purity: The OD 260/280 ratio should fall within the range of 1.7 to 1.9.
Total Sample Amount: It is recommended that the total amount of each sample is not less than 50 ng.
Sample Solvent: Samples can be dissolved in ddH2O or TE buffer (pH 8.0).
Bacterial Whole Genome Sequencing Service
Bacteria Genome Sequencing Finish Map
Combining the advantages of next generation sequencing (NGS) and third generation sequencing platform, we present a fully confirmed bacterial whole-genome assembly. The Nanopore platform provides long read sequences, yielding superior results in genome assembly, structural variation detection, and identification of continuous repetitive sequences. This is complemented by the high coverage depth and precise sequencing quality of Illumina NGS data, enabling the accurate assembly of a comprehensive and precise genome map.
The report includes quality control (QC) of samples and data, sequence assembly and alignment (using BlastN, BlastX), gene annotation, database analyses (GO, COG, KEGG), and a Genome map. Additionally, we provide data for uploading and registering in the NCBI genome database, along with information relevant to genome patent registration.
By leveraging the strengths of both NGS and Nanopore third generation sequencing platforms, we ensure a 100% confirmed bacterial whole-genome assembly. The long read sequences from Nanopore facilitate superior genome assembly, especially in discerning structural variations and identifying continuous repetitive sequences. When combined with the high coverage depth and precise sequencing quality of Illumina NGS data, our approach results in the accurate assembly of a comprehensive and precise whole-genome map.
The report encompasses QC of samples and data, sequence assembly and alignment using tools such as BlastN and BlastX, gene annotation, database analyses including GO, COG, and KEGG, as well as a Genome map. Moreover, it provides the necessary data for uploading to and registering in the NCBI genome database, along with information pertinent to genome patent registration.
Sample Requirements :
Sample Purity: OD 260/280 ratio should be between 1.7 and 1.9.
Sample Concentration: Minimum concentration should not be less than 100 ng/µl.
Total Sample Amount: The recommended total amount is 5 µg, with a minimum of at least 1 µg.
Sample Solvent: Samples can be dissolved in ddH2O or TE buffer (pH 8.0).

Strain Identification
Advantages of Strain Identification Service :
1. Genetic identification methods analyze bacterial 16S and fungal ITS, capable of identifying at the "species" level.
2. Both agar plates and liquid cultures of individual strains are applicable. Genomic DNA of microorganisms can also be submitted for identification.
3. Reports are generated within 5 working days upon sample receipt.
4. A dedicated data management system is available for customers to download files.